Structural biology, biochemistry, and physiology of neuron/cancer signaling
Overview
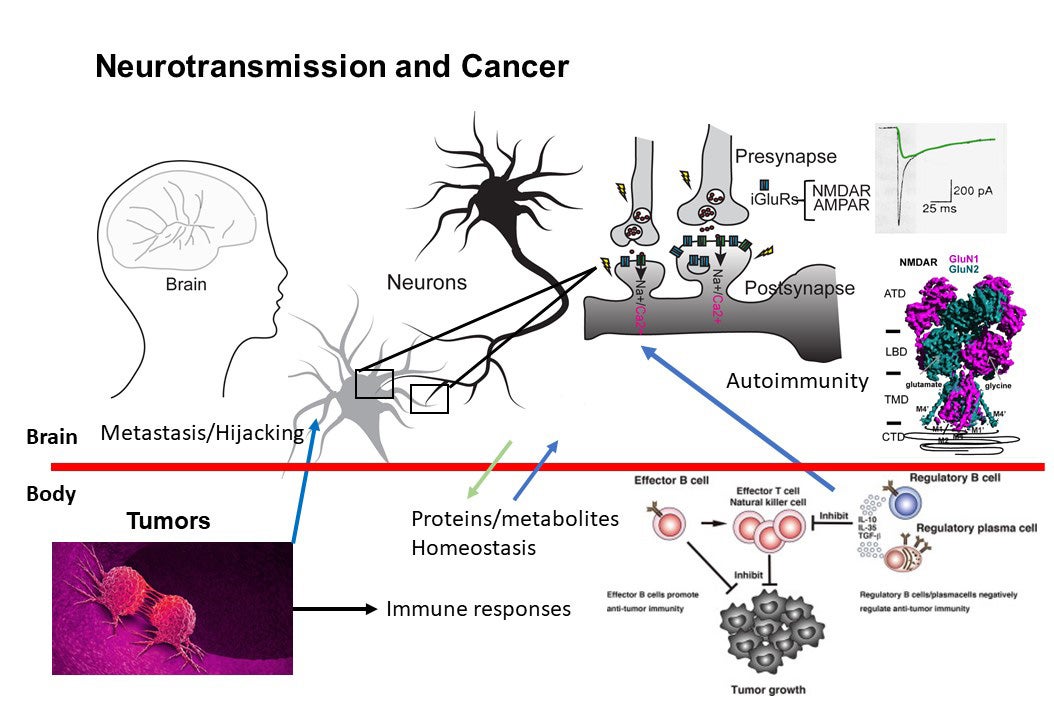
Higher-order brain functions, including learning and memory formation, result from cellular signal transduction processes driven by the assembly of macromolecules in neurons that respond to environmental stimuli. Dysfunction within this macromolecular machinery is often linked to neurological disorders such as schizophrenia, epilepsy, stroke, depression, Parkinson’s disease, and Alzheimer’s disease—conditions that continue to present significant and debilitating clinical challenges. Our broad research interests focus on critical cellular paradigms in neuroscience, such as neuroplasticity and neurodegeneration, which are mediated by alterations in membrane potentials and dynamic protein-protein interactions. Recently, we have also expanded our scope to explore the biology at the interface between neurons and cancer in the context of autoimmunity. Our research encompasses (1) the biochemistry and structural biology of membrane-embedded or membrane-associated receptors and their interacting signaling molecules by cryo-EM and x-ray crystallography, (2) functional analyses of ion channels through electrophysiological techniques, (3) protein engineering for the development of reagents for therapy, detection, and diagnosis, (4) experimental and computational approaches for compound development, (5) imaging of protein complexes using both light and electron microscopy, and (6) understanding interactions between cancers and neurons in the context of autoimmune disease.
Examples of techniques we implement
Research in the Furukawa lab integrates a multidisciplinary approach to address fundamental biological questions. Our methodologies span structural biology, electrophysiology, biophysics, cell biology, protein engineering, and small molecule development. Key structural techniques include X-ray crystallography and single-particle electron-cryo microscopy (cryo-EM). Additionally, we utilize molecular dynamics (MD) simulations to complement and refine our experimental findings in structural biology and pharmacology. We employ a range of electrophysiological techniques, such as patch-clamp and two-electrode voltage-clamp, to detect and analyze the electrical signals generated by ion channels and transporters. The binding affinities of compound-protein and protein-protein interactions are measured using advanced biophysical techniques, including isothermal titration calorimetry (ITC), surface plasmon resonance (SPR), and microscale thermophoresis. Collaborative work with CSHL’s proteomics facility allows us to identify novel protein-protein interactions within cells. Our synthetic biology efforts, including yeast and phage display and directed protein evolution, facilitate the engineering and production of proteins such as antibodies and proteases. We also collaborate with chemists at CSHL to facilitate our chemical-biological efforts. To support our research, we frequently develop and optimize new methodologies; for instance, we recently devised the EarlyBac system, which identifies the optimal combination of UTRs and promoters for the efficient expression and assembly of multimeric membrane protein complexes. Hypotheses generated from any of these disciplines are cross-validated using complementary techniques. For example, structural insights can be tested through electrophysiology and biophysical analyses, and findings from these approaches can inform further structural investigations. Below, we provide an overview of some of the current projects underway in our lab.
Structure and Function of intact NMDA receptor ion channels
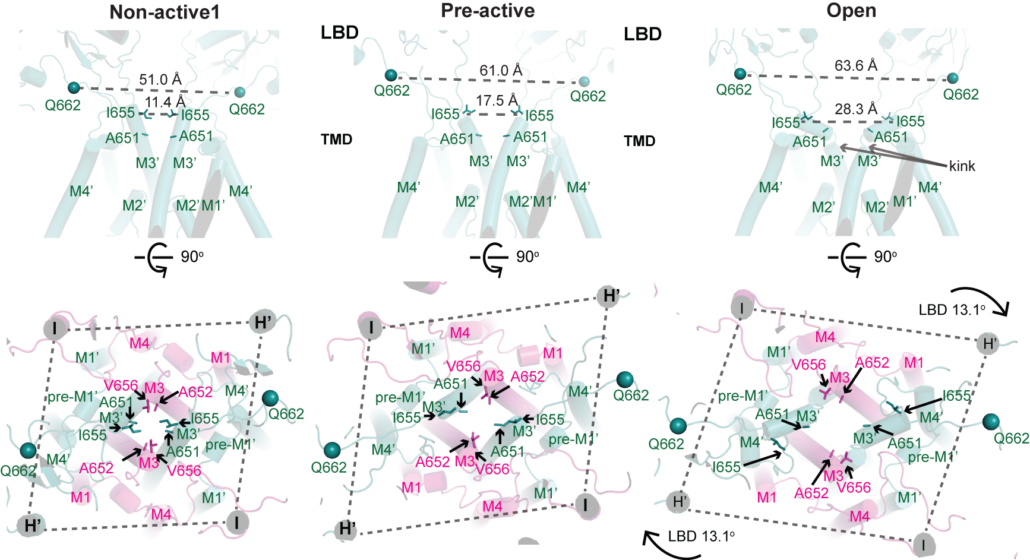
NMDA receptors are heterotetrameric cationic ion channels that are composed of two GluN1 subunits and two GluN2 (A-D) and/or GluN3 (A-B) subunits. GluN1 and GluN3 subunits bind glycine whereas GluN2 subunits bind the neurotransmitter, glutamate. Concurrent binding of glycine and glutamate is necessary for the opening of the cationic ion channel, which contributes to the generation of excitatory postsynaptic potential (epsp). Furthermore, voltage-dependent calcium influx by NMDA receptors drives cellular signaling for neuroplasticity, which is critical for learning and memory formation. Dysfunctional NMDA receptors are implicated in Alzheimer’s disease, Parkinson’s disease, depression, schizophrenia, stroke, and autism. We solved the crystal structure of the intact heterotetrameric GluN1-GluN2B NMDA receptors from rat in complex with glycine, glutamate, and an allosteric inhibitor, ifenprodil. This structure revealed how the extracellular domains, amino-terminal domains (ATDs), and ligand-binding domains (LBDs), from both GluN1 and GluN2 subunits are associated with each other in the ‘dimer of GluN1-GluN2 heterodimers’ manner. By a combination of x-ray crystallography and single-particle electron cryo-microscopy (cryo-EM), we attempt to understand the regulatory mechanisms of NMDA receptors. Specifically, we are working toward understanding how multiple domains (ATD, LBD, and TMD) move with respect to one another in various functional states. The state-of-the-art cryo-EM facility in our building allows us to explore the above at high quality. In combination with electrophysiology and MD simulation, we have been addressing functional questions including allosteric modulations, pH-sensitivity mediated by alternative splicing, activation, and competitive inhibition.
Structure and reagent of NMDA receptors
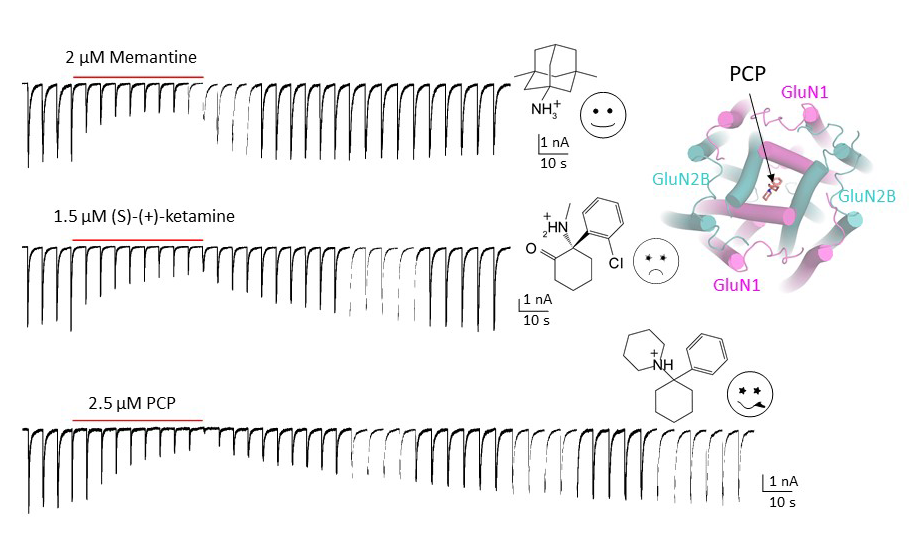
Since the discovery of NMDA receptors in the early 1970s, considerable efforts have focused on characterizing NMDA receptor pharmacology. NMDA receptors exhibit significant diversity: eight splicing variants of the GluN1 subunit can assemble as heterotetramers with four types of GluN2 (A-D) and two types of GluN3 (A-B), forming ion channels with distinct functional properties, including varied open probabilities, opening/closing kinetics, and compound binding affinities and potencies. Subtype-specific compounds targeting NMDA receptors hold promise for treating neurological diseases and disorders, making a high-resolution understanding of the molecular determinants of subtype specificity critical. To this end, we use cryo-EM to determine the structures of intact heterotetrameric NMDA receptors, particularly for studying compounds such as channel blockers (e.g., ketamine, PCP, memantine) that interact with the transmembrane domain (TMD). We employ X-ray crystallography for compounds that bind exclusively to the extracellular domains (ATD or LBD). By resolving these domains in complex with target compounds to high-resolution levels (2.0 Å and beyond), we can accurately visualize drug-binding sites, critical water molecules, and conformational changes within these domains. Our objective is to provide structural insights that allow for precisely mapping drug-binding sites at atomic resolution, advancing the design of targeted therapeutic compounds in pharmacology. We also aim to identify previously uncharted compound binding sites to uncover novel mechanisms of channel regulation.
Deciphering anti-NMDA receptor encephalitis
Anti-NMDAR encephalitis is the most prevalent form of immune-mediated encephalitis, predominantly affecting young individuals, typically in their second and third decades of life. Although the NMDAR has been clearly identified as the autoimmune target, the disease’s precise mechanisms remain incompletely understood. This gap in knowledge poses significant challenges and is further exacerbated by suboptimal diagnostic methods. Early diagnosis and targeted treatment are essential, as delays in intervention can lead to worsening clinical outcomes, including psychiatric disturbances such as psychosis and behavioral changes, as well as seizures and central hypoventilation. Anti-NMDAR encephalitis is often linked to ovarian teratomas and other cancers that express NMDARs. We examined three IgGs isolated from three patients and demonstrated that they bind to distinct epitopes within NMDAR tetramers. Additionally, we found that these antibodies exhibit varied stoichiometry when binding to the NMDAR tetramers. Our electrophysiological studies revealed inhibition of spontaneous excitatory postsynaptic currents (EPSCs) in cultured neurons. Our findings underscore the epitope diversity and structural and functional heterogeneity of anti-NMDAR antibodies among patients. The Furukawa lab now possesses a robust and effective workflow for testing antibodies isolated from human patients or animal models. This positions us uniquely to elucidate the molecular mechanisms underpinning anti-NMDAR encephalitis and potentially inform new therapeutic strategies.
Large-pore channels
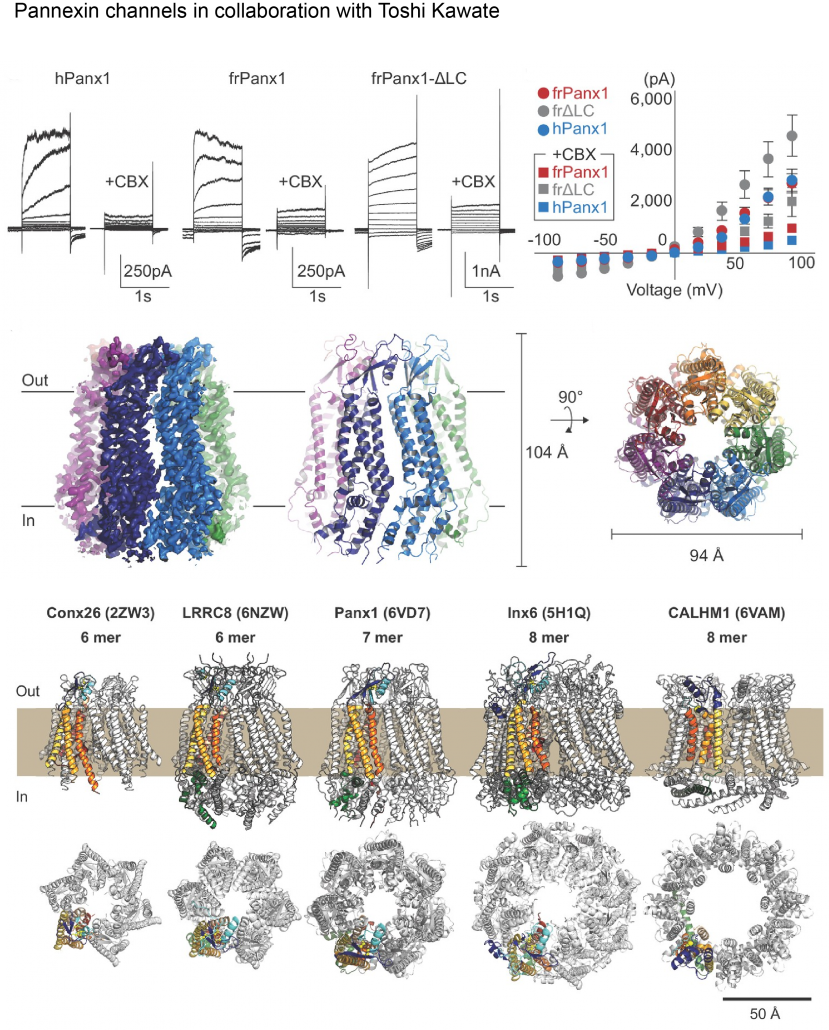
Biological membranes in various tissues and organs contain large-pore channels that facilitate the permeation of diverse ions and metabolites, such as ATP. Notable examples include the connexin, innexin, pannexin, and LRRC8 channel families, which form gap junctions and functional cell surface channels. These channels utilize multiple regulatory mechanisms, such as voltage-gating, to maintain cellular homeostasis and viability, supporting various biological functions. Among the most recently discovered large-pore channels are the calcium homeostasis modulators (CALHMs), which enable the voltage-dependent passage of small ions and ATP, playing key roles in physiological processes like neuronal excitability, taste signaling, and conditions such as depression and Alzheimer’s disease. Despite their critical functions, the structural details and patterns of oligomeric assembly of these channels remain unclear. To address this, we use cryo-EM and electrophysiological to study how CALHM proteins assemble and facilitate voltage-dependent ion and metabolite permeation. Additionally, we investigate the structure and function of another large-pore channel, Pannexin, known for its role in ATP efflux.
Structural pharmacology of cancer-related membrane proteins
The cancer biology community at CSHL is known for its highly interactive and collaborative nature. Leveraging our expertise, we are beginning to investigate membrane proteins that play critical roles in cancer cell survival—proteins that can serve as significant vulnerabilities when targeted effectively. Our aim is to elucidate their biochemical properties, structural characteristics, and strategies for therapeutic targeting. Through close collaboration with the Cancer Center at CSHL, the Furukawa lab is set to deepen its engagement in cancer biology.
Reagent development – chemical biological, synthetic biological, and antibody engineering approach
Antibodies are extremely useful biological reagents that can recognize and functionally regulate target proteins. We are screening and developing antibodies for cell surface receptors expressed in neurons and cancer cells. This is done by basic immunization technology and/or the phage- and yeast-display methods. Identified antibodies are further tested for functional effects and are re-engineered using structural information.We further combine this synthetic biological approach with chemical biology in collaboration with John Moses’s lab at CSHL. We are developing reagents that can be used as therapeutic reagents, biomarkers, and tools for basic researches.